Szabó
Laboratory
Developmental Reprogramming
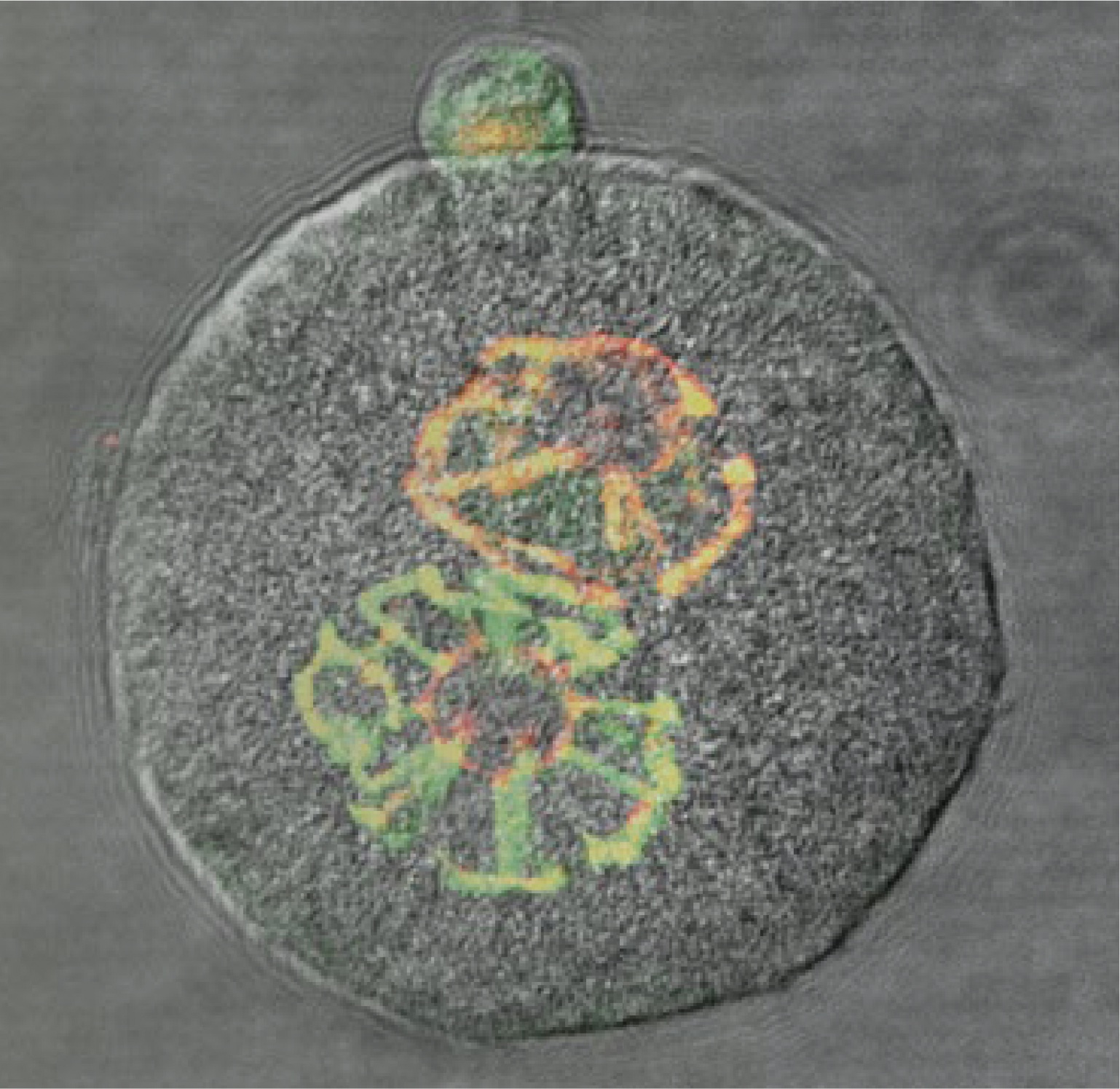
Both the soma-germline and the germline-soma transitions involve global erasure and reestablishment of DNA methylation patterns. At the soma-germline transition, the paternally and maternally inherited sets of chromosomes are prepared separately in the male and in the female germlines. The chromosomes in the sperm and egg contribute to the next generation when they join at fertilization. Soon after fertilization––at the germline-soma transition––the two half genomes undergo another wave of global remodeling initiating somatic development. The chromosomes inherited from the sperm or the egg carry with them into the soma an epigenetic memory of the male or female germlines, which is detectable in the parental-allele-specific transcription of imprinted genes.
The Szabó Laboratory is interested in the mechanisms of how DNA methylation is erased in primordial germ cells and in the zygote. We also study the patterning of de novo DNA methylation in fetal male germ cells. We use the tools of molecular biology and mouse genetics to map the changes in the epigenome and identify the specific molecules that take part in these global and imprinted locus-specific processes. In addition, we test whether the natural epigenetic reprogramming processes of the germline are sensitive to environmental insults, potentially leading to transgenerational epigenetic inheritance.
Join Our Team!
The lab of Dr. Piroska E. Szabó is looking for a skilled and collaborative postdoctoral fellow to study mechanisms relevant for the biological processes that remodel the epigenome between generations. The lab focuses on biological processes that take place in the germ line and the embryo, such as genomic imprinting, maternal effects of histone methyltransferases, and the remodeling of Z-DNA structure.
Recent News & Featured Publications
Learn More
Study illuminates mechanism that annotates genetic information passed from fathers to offspring
Early study points to potential therapeutic avenue for a pair of rare pediatric diseases

Meet the scientist behind the science: Dr. Piroska Szabó
Liao J, Song S, Gusscott S, Fu Z, Vanderkolk I, Busscher BM, Lau KH, Brind’Amour J, Szabó PE. 2023. Establishment of paternal methylation imprint at the H19/Igf2 imprinting control region. Sci Adv 9(36).
Meng Y, Wang G, He H, Lau KH, Hurt A, Bixler BJ, Parham A, Jin SG, Xu X, Vasquez KM, Pfeifer GP, Szabó PE. 2022. Z-DNA is remodelled by ZBTB43 in prospermatagonia to safeguard the germline and epigenome. Nat Cell Biol 24(7): 1141–1153.
Zeng TB, Pierce N, Liao J, Singh P, Lau K, Zhou W, Szabó PE. 2021. EHMT2 suppresses the variation of transcriptional switches in the mouse embryo. PLOS Genet.
Liao J, Zeng TB, Pierce N, Tran DA, Singh P, Mann JR, Szabó PE. 2021. Prenatal correction of IGF2 to rescue the growth phenotypes in mouse models of Beckwith-Wiedemann and Silver-Russell syndromes. Cell Rep 34(6):108729.
Zeng, TB, Pierce N, Szabó PE. 2021. H3K9 methyltransferase EHMT2/G9a controls ERVK-driven non-canonical imprinted genes. Epigenomics 13(16).
Zeng TB, Han L, Pierce N, Pfeifer GP, Szabó PE. 2019. EHMT2 and SETDB1 protect the maternal pronucleus from 5mC oxidation. Proc Natl Acad Sci U S A 116(22):10834–10841.
Our Impact
We’re raising thousands to save millions.
We’re turning hope into action for the millions of people around the world affected by diseases like cancer and Parkinson’s. Find out how you can help us make a difference.
- 122 peer-reviewed papers published in 2024, 63 of which were in high-impact journals
- 15 VAI-SU2C Epigenetics Dream Team clinical trials launched to date
- 10 clinical trials co-funded by VAI & Cure Parkinson's (out of 41 total International Linked Clinical Trials Program trials)
Piroska Szabó, Ph.D.
Associate Professor, Department of Epigenetics
Areas of Expertise
Epigenetics, genomic imprinting, insulator, developmental reprogramming, germ line, DNA methylation, chromatin
Biography
Dr. Piroska Szabó earned an M.Sc. in biology and a Ph.D. in molecular biology from József Attila University, Szeged, Hungary. She joined Beckman Research Institute of City of Hope, Duarte, Calif. in 1992 as a postdoctoral fellow. She served first as an assistant research scientist and associate research scientist before becoming an assistant professor in the Department of Molecular and Cellular Biology at Beckman Research Institute in 2006 and was promoted to associate professor in 2011. She joined VAI in 2014 as an associate professor in the Department of Epigenetics.
Background and past work
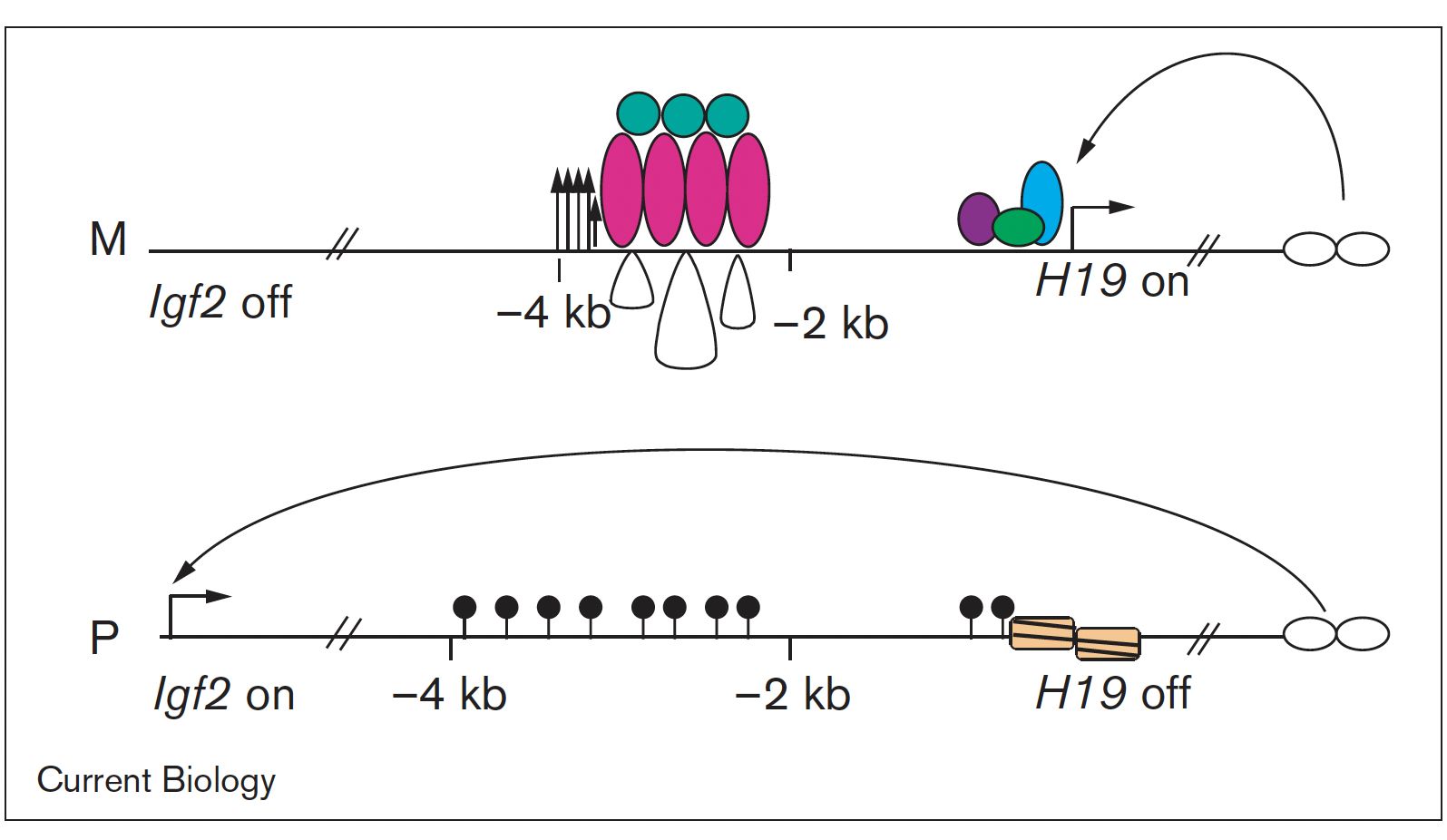
Imprinted genes are unusual because their activity states depend on parental inheritance of the chromosomes. The paternal or maternal origin of chromosomes is established in the male and female germ lines, respectively. The imprint mark is most often DNA methylation at germ line differentially methylated regions (DMRs), which is then specific to the sperm or the oocyte. One interesting question was the mechanism of how the soma interprets the male gamete-specific DNA methylation imprint at the H19/Igf2 imprinted domain. We showed using in vivo footprinting and ligation-mediated PCR, that the imprinting control region works as a methylation-sensitive allele-specific CTCF-binding insulator (Szabó et al., Curr Biol, 2000). CTCF insulates the Igf2 promoter from the enhancers in the maternally inherited chromosome, but due to DNA methylation CTCF cannot bind at the same sequences in the paternal chromosome, thus Igf2 is not insulated from the enhancer, and it is expressed there.
We later confirmed this model using genetically altered mice (for review, see Singh P and Szabó P, Front Genet, 2012).
Prenatal correction of IGF2 to rescue the growth phenotypes in mouse models of Beckwith-Wiedemann and Silver-Russell syndromes
The insulator model of imprinting gives explanation for specific cases of two human imprinting disorders. Beckwith-Wiedemann syndrome (BWS) and Silver-Russell syndrome (SRS) are childhood diseases, which manifest in fetal overgrowth and growth retardation, respectively. The growth anomalies underlie many complications after birth, such as the predisposition to childhood cancers, implying that correcting growth anomalies may prevent subsequent complications. The growth symptoms in a large subset of BWS and SRS cases are consistent with genetic or epigenetic molecular anomalies of one interesting chromosomal region, conserved in humans and mice.
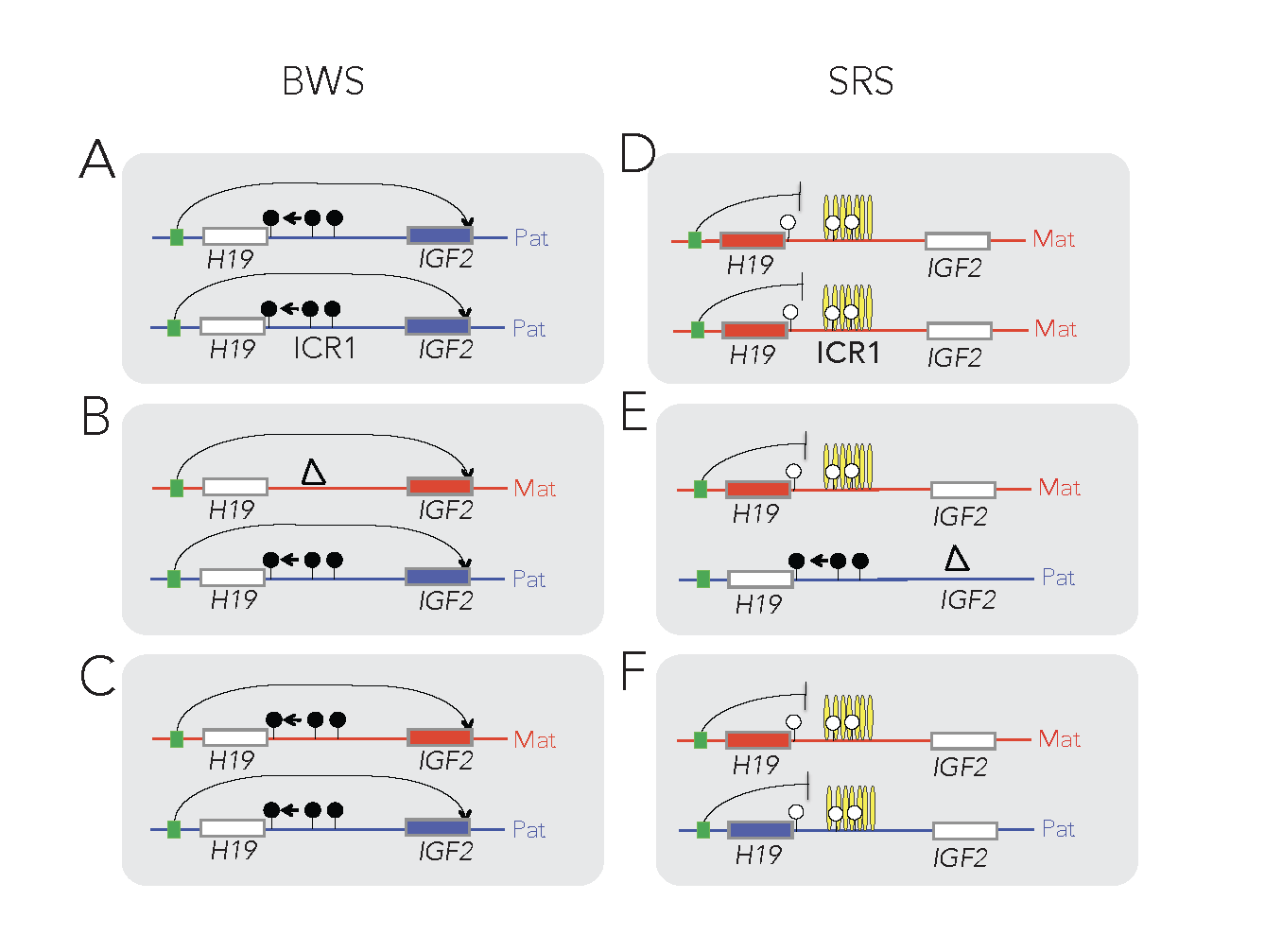
This region harbors imprinted genes, including the paternally expressed insulin-like growth factor 2 (IGF2) gene, encoding IGF2 protein, a key facilitator of fetal and placental growth. IGF2 overexpression is consistent with BWS molecular aberrations and fetal overgrowth, and the absence of IGF2 is consistent with SRS molecular features and fetal growth retardation. One clinical challenge is that IGF2 is not targeted in BWS or SRS. The other clinical challenge is that growth regulation is disturbed in BWS and SRS before birth, but medical interventions manage the symptoms of these diseases only after birth. Importantly, Igf2 mRNA levels plummet after birth, which implies that any intervention that targets IGF2 has to occur prenatally. We recently carried out a set of mouse experiments to find novel approaches for the diagnosing and prevention of BWS and SRS in human children (Liao et al., Cell Rep, 2021).
Using genetic and pharmacological approaches and the gene-targeted mouse lines that I developed earlier, we found that IGF2-dependent BWS and SRS cases can be identified, and that IGF2 action can be normalized prenatally to prevent growth aberrations and their complications. The results collectively provide key mechanistic data for future human studies aimed at intervention in IGF2- dependent BWS and SRS cases.
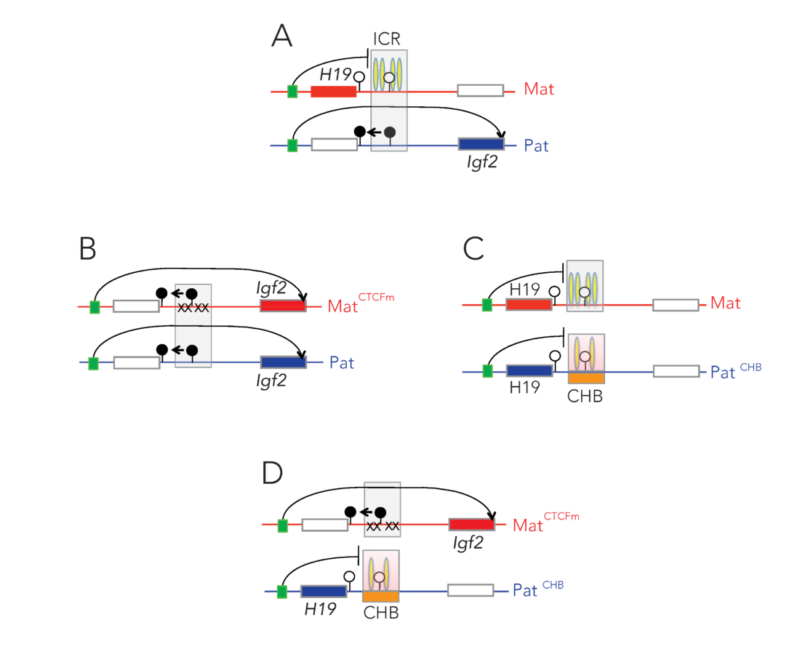
Noncanonical imprinting
Recently, a new type of genomic imprinting was described, called non-canonical imprinting. These imprinted genes do not depend on germline DMRs, but rather on the presence of the Polycomb mark, H3K27me3, in the oocyte. Non-canonical imprinted genes are under the control of ERVK repeats. We tested the role of an H3K9 dimethyltransferase, G9a/EHMT2 in non-canonical imprinting using our mouse knockout model and allele-specific RNA sequencing. We found that EHMT2 is required for maintaining the paternal-allele-specific expression by suppressing the maternal alleles of the ERVK promoters in the ectoplacental cone. Analyzing embryos, we found that EHMT2 is required for suppressing both parental alleles in the embryo by targeting DNA methylation to the ERVK promoters. Our study identifies EHMT2 as a novel and essential player in the non-canonical imprinting mechanism (Zeng TB, Pierce N & Szabó PE, Epigenomics, 2021).
Background and past work
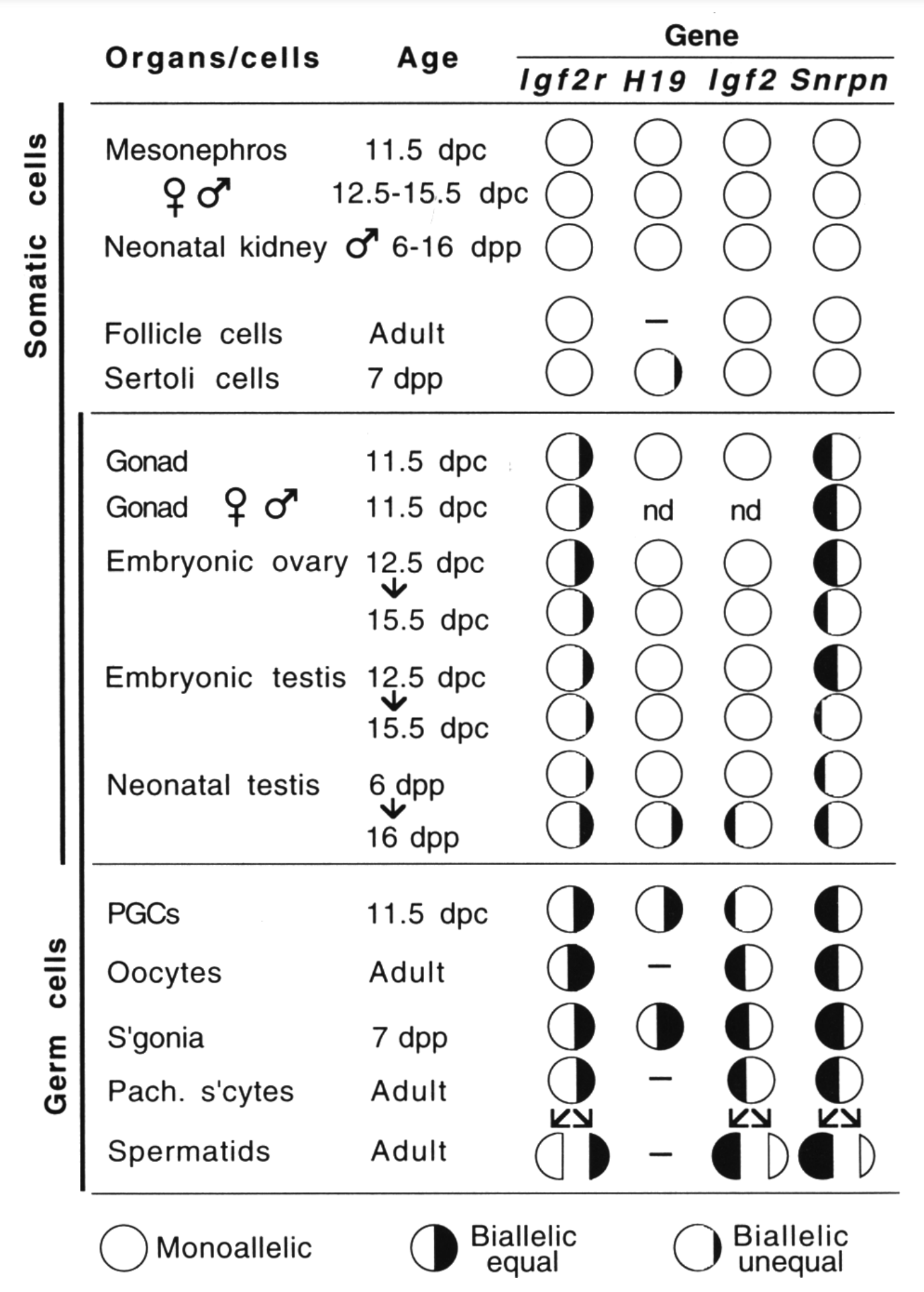
By analyzing allele-specific transcription in embryonic and fetal germ cells earlier, we showed that imprinted expression is neutralized in the germ line (Szabó et al., Genes & Dev 1995). This discovery opened up a new direction toward understanding the mechanism that resets the imprints in the male and female germ lines.
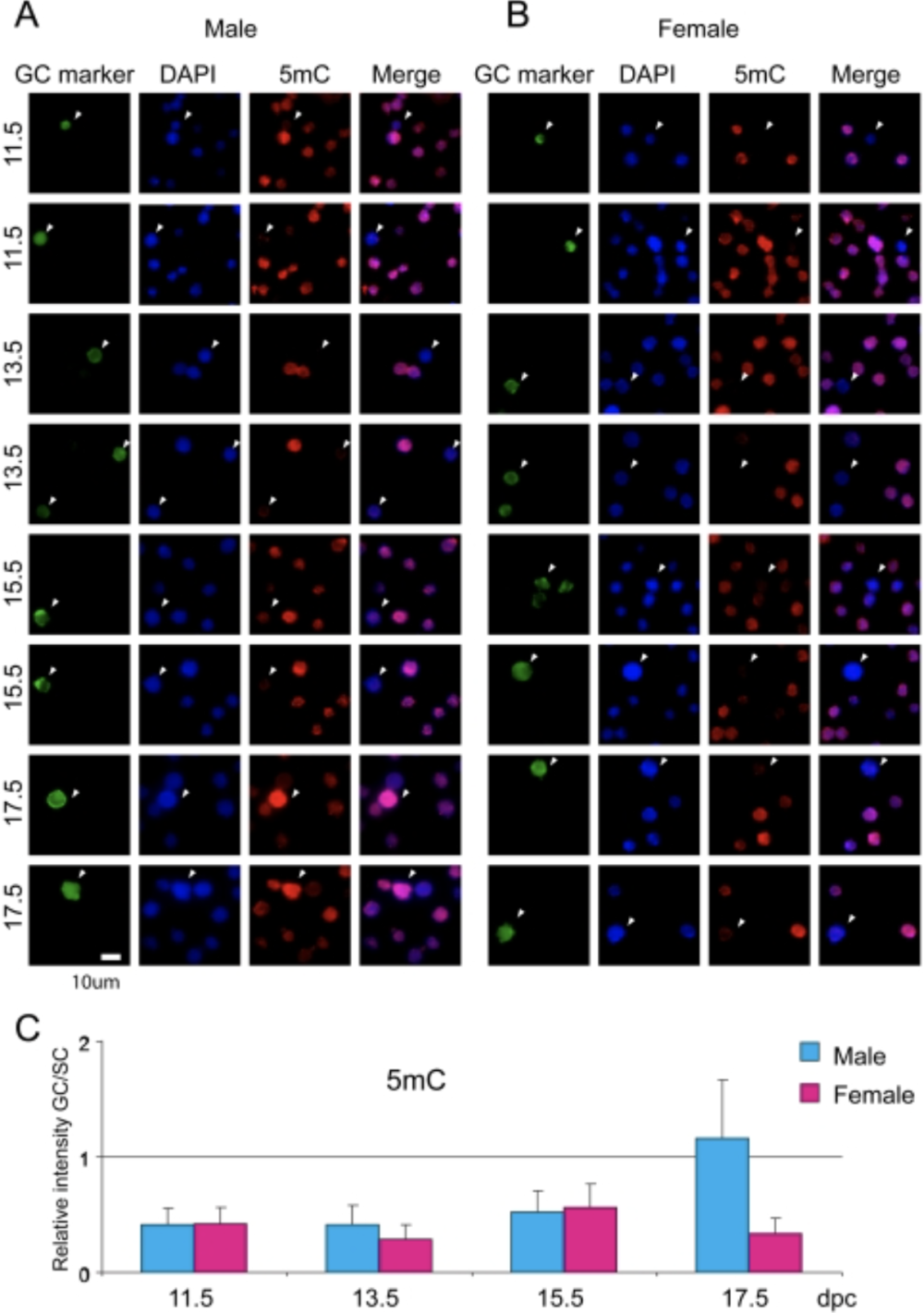
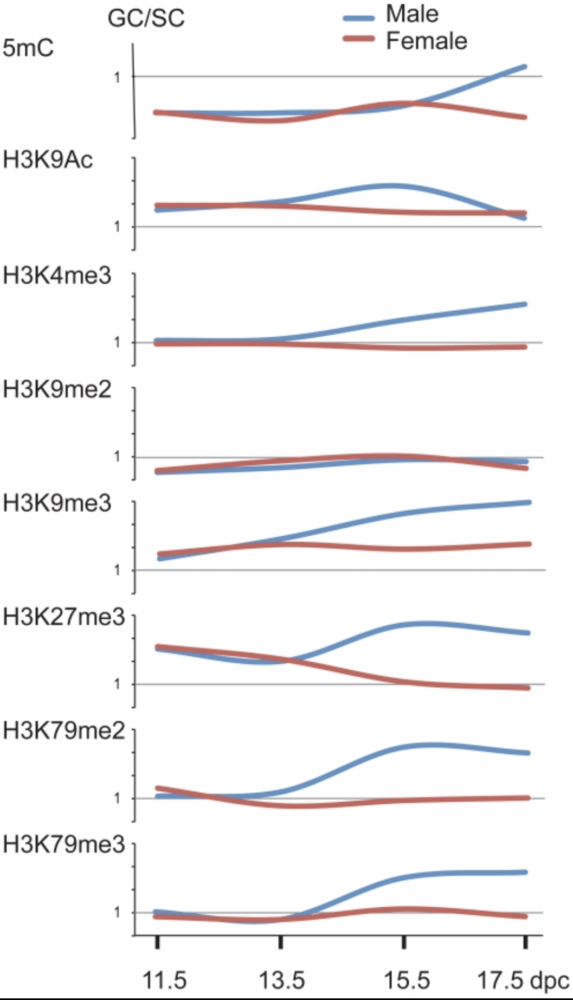
Sex-specific dynamics of global chromatin changes in fetal mouse germ cells
Mammalian germ cells undergo global reprogramming of DNA methylation during their development. Global DNA demethylation occurs around the time when the primordial germ cells colonize the embryonic gonads, and this coincides with dynamic changes in chromatin composition. Global de novo DNA methylation takes place with remarkably different dynamics between the two sexes: prospermatogonia attaining methylation during fetal stages and oocytes attaining methylation postnatally. Our hypothesis was that dynamic changes in chromatin composition may precede or accompany the wave of global DNA de novo methylation as well. We used immunocytochemistry to measure global DNA methylation and chromatin components in male and female mouse fetal germ cells compared to control somatic cells of the gonad. We found that global DNA methylation levels sharply increased in male germ cells at 17.5 days post coitum but remained low in female germ cells at all fetal stages. Global changes in chromatin composition: i. preceded global DNA methylation in fetal germ cells; ii. sex specifically occurred in male but not in female germ cells; iii. affected active and repressive histone marks; and iv. included histone tail and histone globular domain modifications. Our data suggest that dynamic changes of chromatin composition may provide a framework for the pattern of male-specific de novo DNA methylation in prospermatogonia (Abe M et al., PLoS One, 2011).
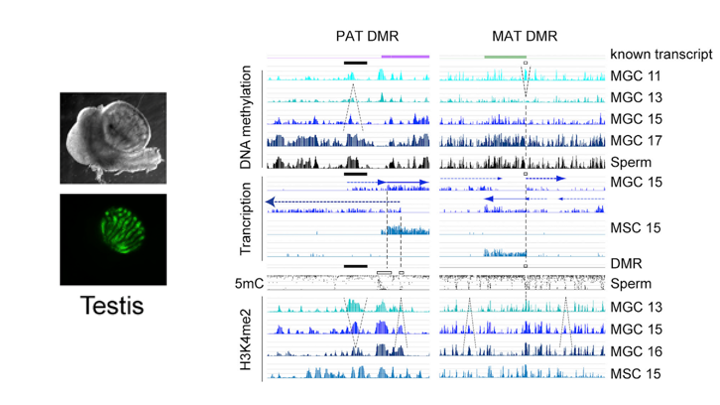
De novo DNA methylation in the male germ line is excluded at sites of H3K4 methylation
To understand what dictates the emerging patterns of de novo DNA methylation in the male germline, we mapped DNA methylation, chromatin and transcription changes in purified fetal mouse germ cells (MGC) by using methylated CpG island recovery assay (MIRA)-chip, chromatin immunoprecipitation (ChIP)-chip, and strand-specific RNA deep sequencing, respectively. Global de novo methylation occurred by default in prospermatogonia without any apparent trigger from preexisting repressive chromatin marks but was preceded by broad, low-level transcription along the chromosomes, including the four paternally imprinted differentially methylated regions (DMRs). Default methylation was excluded only at precisely aligned constitutive or emerging peaks of H3K4me2, including most CpG islands and some intracisternal A particles (IAPs). Similarly, each maternally imprinted DMR was protected from default DNA methylation among highly methylated DNA by an H3K4me2 peak and transcription initiation at least in one strand. Our results suggest that the pattern of de novo DNA methylation in prospermatogonia is dictated by opposing actions of broad, low-level transcription and dynamic patterns of active chromatin. (Singh et al., Cell Rep, 2013).
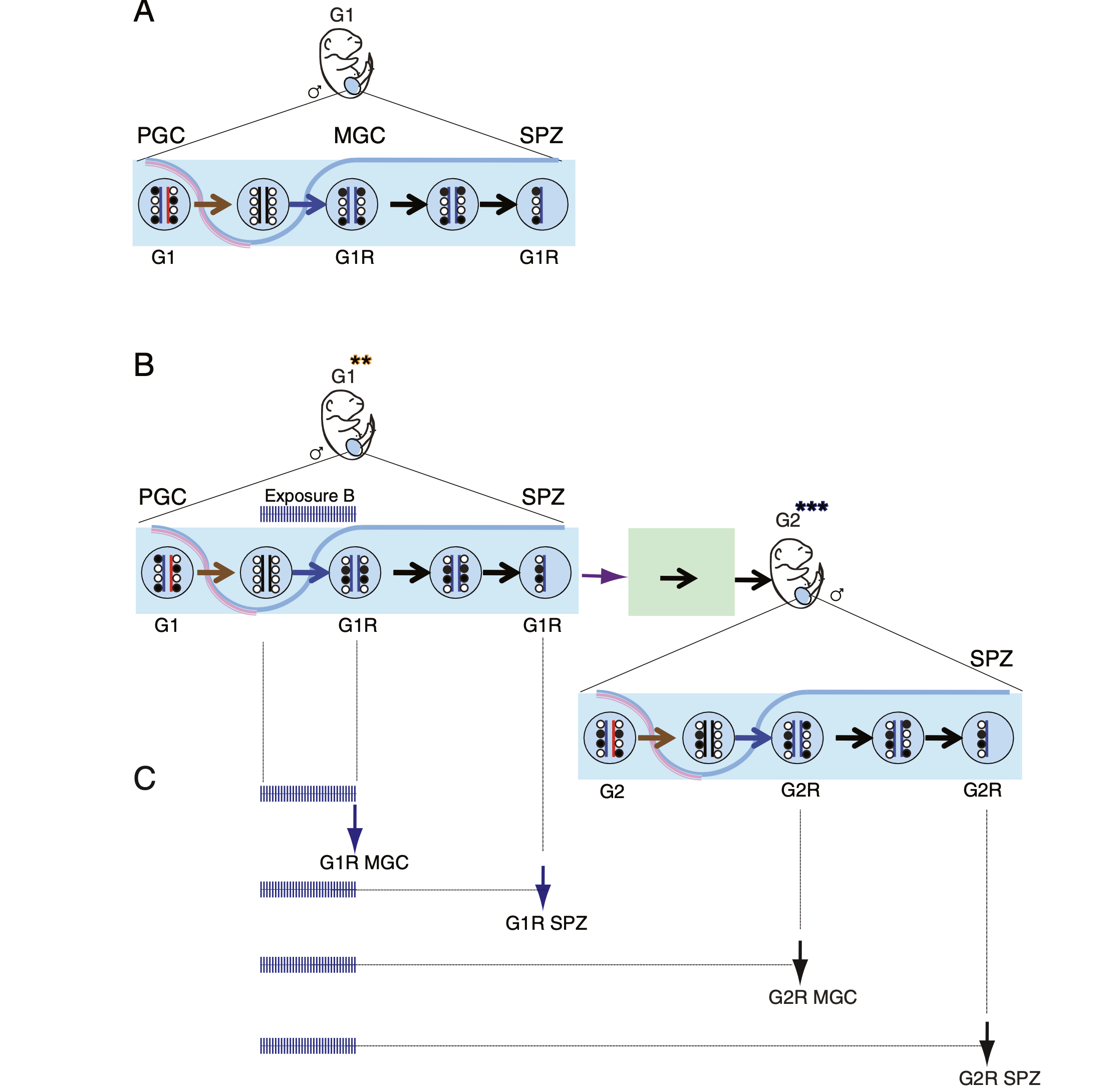
Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming
The success of our genome-wide mapping studies in the germ line (Singh et al., Cell Rep, 2013) prompted us to ask whether the remodeling events can be disturbed by environmental insults, such as endocrine disruptors, which had been implicated in transgenerational epigenetic inheritance. We found that such aberrations arise in fetal germ cells but are later effectively repaired in the germ line of the next generation (Iqbal et al. Genome Biol, 2015).
Understanding the drivers of early epigenetic events bears high significance in reproductive medicine. Our long-time interest is to understand the role of histone methyltransferases in embryo development. It is important to genetically distinguish the maternal effect of an epigenetic modifier from its zygotic effect. Parental mutations, even without being inherited to the offspring, may result in a phenotypic change, such as growth retardation. By examining maternal-effect mutants, we can gain insight into problems in fecundity (which may be misdiagnosed as infertility), genetically unexplained losses of pregnancy, and delayed, growth-retarded fetal development.
Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine
Shortly after fertilization, global epigenome remodeling takes place. Strikingly, the paternal pronucleus of the zygote undergoes a rapid active DNA demethylation whereas the maternal pronucleus is demethylated passively. We showed that shortly after fertilization 5hmC is created by genome-wide oxidation of 5mC in the paternal genome (Iqbal et al., PNAS, 2011). A subsequent collaboration with Guoliang Xu revealed that TET3 mediates the oxidation of 5mC in the paternal genome (Gu et al., Nature, 2011).
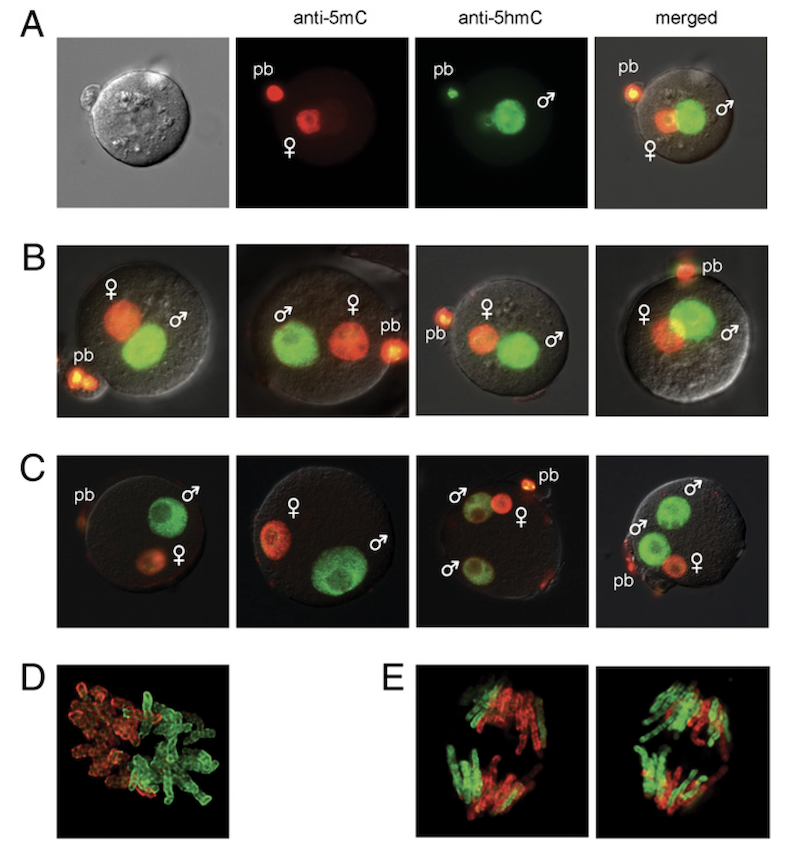
EHMT2 and SETDB1 protect the maternal pronucleus from 5mC oxidation
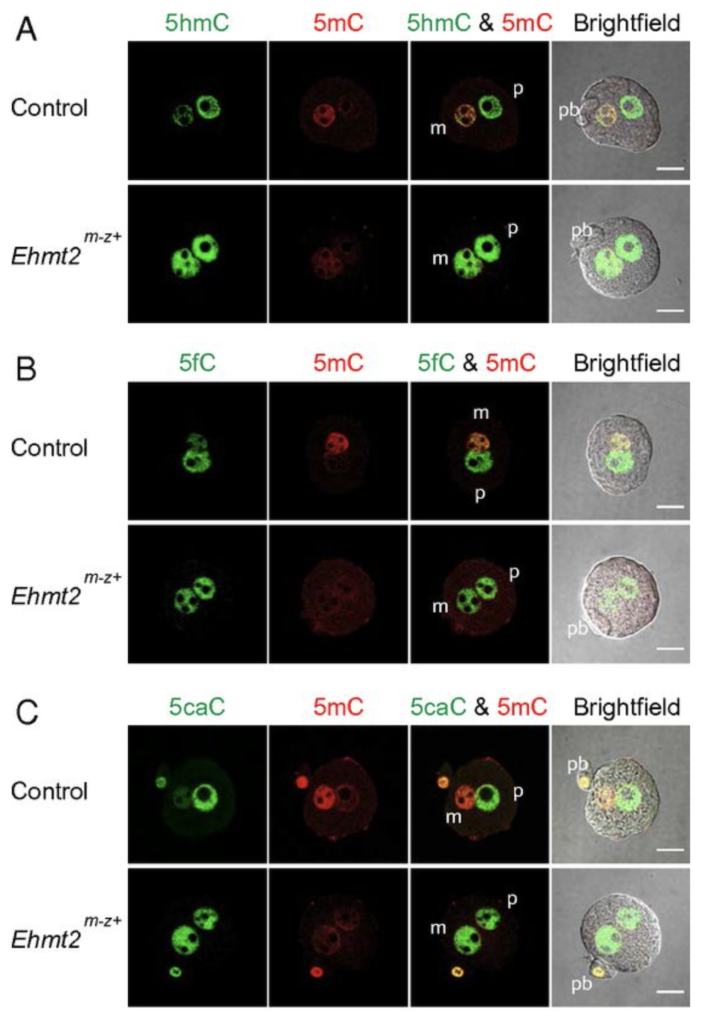
We next addressed the question, what makes the maternal pronucleus in the zygote resistant to the oxidation by TET3. We showed by immunocytochemistry in a maternal knockout experiment that the zygote’s maternal pronucleus is protected from TET3-mediated oxidation by the EHMT2 and SETDB1 proteins, which are H3K9 histone methyltransferases and act as maternal factors (Zeng et al., PNAS, 2019).
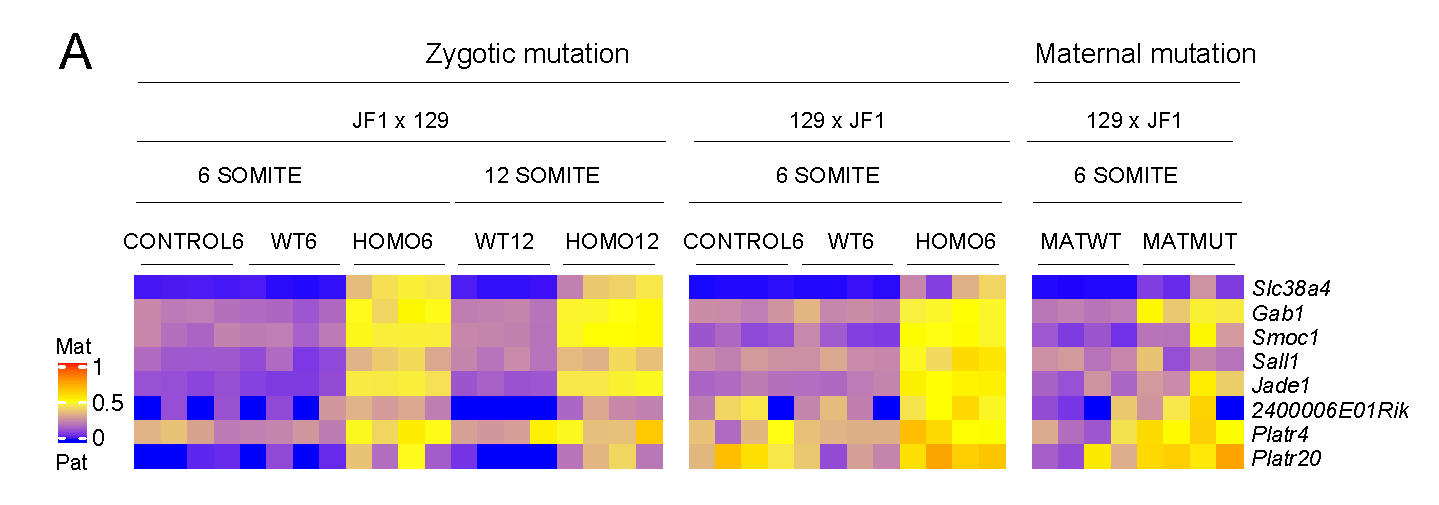
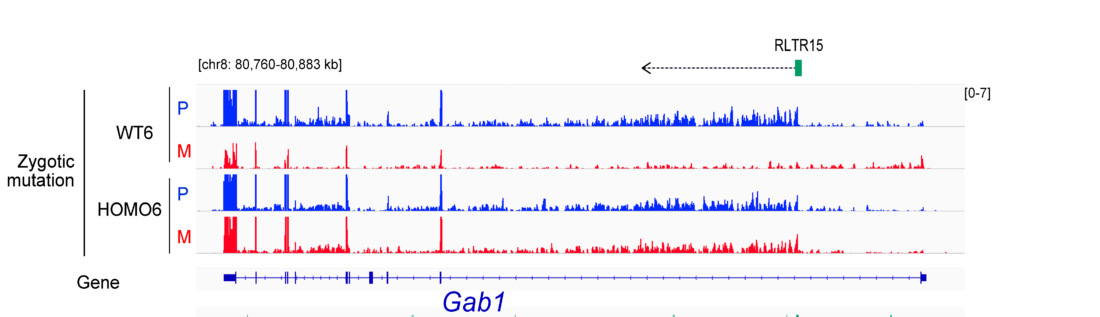
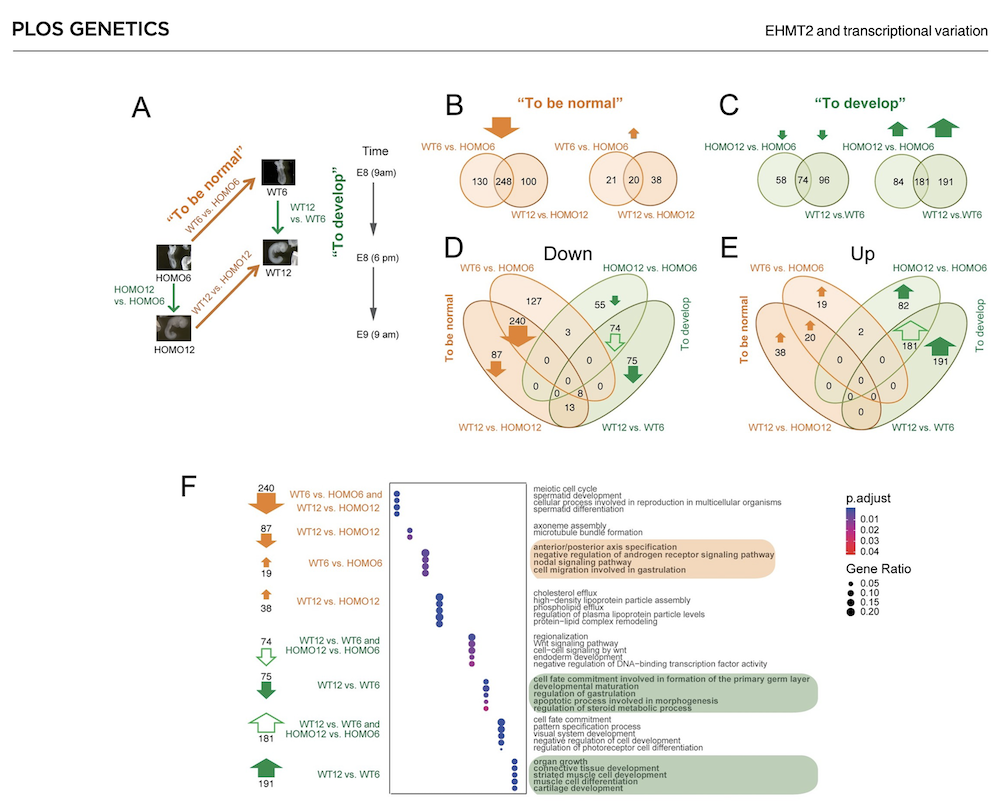
EHMT2 suppresses the variation of transcriptional switches in the mouse embryo
EHMT2 is the main euchromatic H3K9 methyltransferase. Embryos with zygotic, or maternal mutation in the Ehmt2 gene exhibit variable developmental delay. To understand how EHMT2 prevents variable developmental delay we performed RNA sequencing of mutant and somite stage-matched normal embryos at 8.5–9.5 days of gestation. Using four-way comparisons between delayed and normal embryos we clarified what it takes to be normal and what it takes to develop. By comparing wild type and zygotic mutant embryos along the same developmental window we identified transcription changes where precise switching during development occurred only in the normal but not in the mutant embryo. At the 6-somite stage, gastrulation-specific genes were not precisely switched off in the Ehmt2-/- zygotic mutant embryos, while genes involved in organ growth, connective tissue development, striated muscle development, muscle differentiation and cartilage development were not precisely switched on. The Ehmt2mat-/+ maternal mutant embryos displayed high transcriptional variation consistent with their variable survival. Global profiling of transposable elements revealed EHMT2 targeted DNA methylation and suppression at LTR repeats, mostly ERVKs. In Ehmt2-/- embryos, transcription over very long distances initiated from such misregulated “driver” ERVK repeats, encompassing a multitude of misexpressed “passenger” repeats. In summary, EHMT2 reduced transcriptional variation of developmental switch genes and developmentally switching repeat elements at the six-somite stage embryos.
(Zeng T-B et al., PLoS Genetics, 2021).
Maternal DOT1L is dispensable for mouse development
A battery of chromatin modifying enzymes play essential roles in remodeling the epigenome in the zygote and cleavage stage embryos, when the maternal genome is the sole contributor. Here we identify an exemption. DOT1L methylates lysine 79 in the globular domain of histone H3 (H3K79). Dot1l is an essential gene, as homozygous null mutant mouse embryos exhibit multiple developmental abnormalities and die before 11.5 days of gestation. To test if maternally deposited DOT1L is required for embryo development, we carried out a conditional Dot1l knockout in growing oocytes using the Zona pellucida 3-Cre (Zp3-Cre) transgenic mice. We found that the resulting maternal mutant Dot1lmat−/+ offspring displayed normal development and fertility, suggesting that the expression of the paternally inherited copy of Dot1l in the embryo is sufficient to support development. In addition, Dot1l maternal deletion did not affect the parental allele-specific expression of imprinted genes, indicating that DOT1L is not needed for imprint establishment in the oocyte or imprint protection in the zygote. In summary, uniquely and as opposed to other histone methyltransferases and histone marks, maternal DOT1L deposition and H3K79 methylation in the zygote and in the preimplantation stage embryo is dispensable for mouse development.
(Liao and Szabó, Sci Rep, 2020).
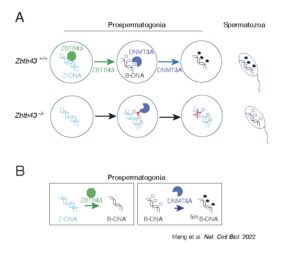
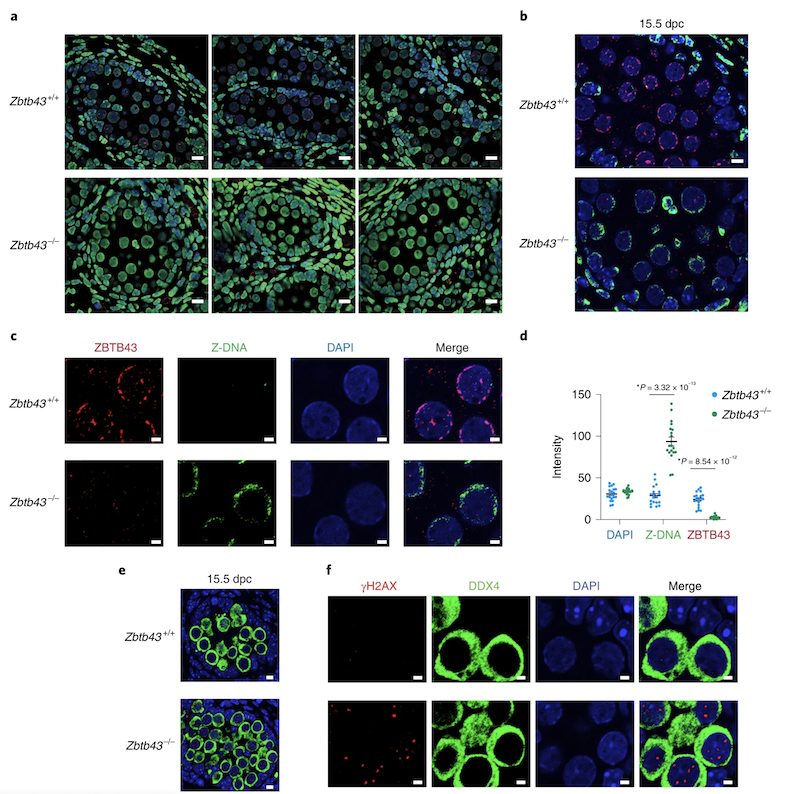
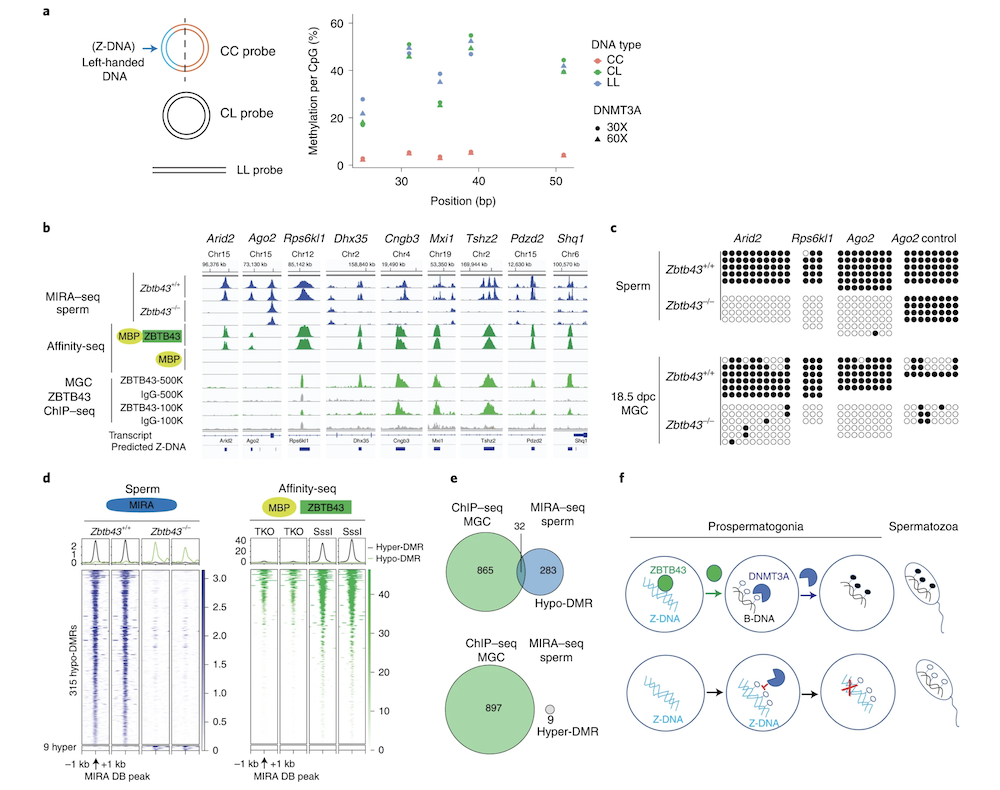
Z-DNA is remodeled by ZBTB43 in prospermatogonia to safeguard the germline genome and epigenome
In addition to the right-handed B-DNA helix, DNA can be detected in alternative forms including a left-handed zigzag shaped double helix, called Z-DNA. Purine–pyrimidine repeats (PPRs), which can adopt Z-DNA conformation and induce DNA double-strand breaks (DSBs), are risk factors for genomic rearrangements and cancer. DNA breaks at potential Z-DNA sites can lead to somatic mutations in cancer or to germline mutations that are transmitted to the next generation. It is not known whether any mechanism exists in the germ line to control Z-DNA structure and DNA breaks at purine–pyrimidine repeats. We found genetic, epigenomic and biochemical evidence for the existence of a novel biological process that erases Z-DNA specifically in germ cells of the mouse male foetus.
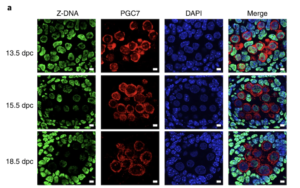
We also identified ZBTB43, a previously uncharacterized zinc finger protein, that binds to and removes Z-DNA. By removing Z-DNA structures, ZBTB43 prevents the formation of DNA double-strand breaks in prospermatogonia. By removing Z-DNA, ZBTB43 also promotes de novo DNA methylation at CG-containing purine–pyrimidine repeats in prospermatogonia. Therefore, the genomic and epigenomic integrity of the species is safeguarded by remodelling DNA structure in the mammalian germ line during a critical window of germline epigenome reprogramming (Meng YY et al., Nature Cell Biology, 2022).
SELECTED PUBLICATIONS
Liao J, Song S, Gusscott S, Fu Z, Vanderkolk I, Busscher BM, Lau KH, Brind’Amour J, Szabó PE. 2023. Establishment of paternal methylation imprint at the H19/Igf2 imprinting control region. Sci Adv 9(36).
Meng Y, Szabo PE. 2022. The remodeling of z-DNA in the mammalian germ line. Biochem Soc Trans 50(6):1875–1884.
Meng Y, Wang G, He H, Lau KH, Hurt A, Bixler BJ, Parham A, Jin SG, Xu X, Vasquez KM, Pfeifer GP, Szabó PE. 2022. Z-DNA is remodelled by ZBTB43 in prospermatagonia to safeguard the germline and epigenome. Nat Cell Biol 24(7): 1141-1153.
Zhou W, Hinoue T, Barnes B, Mitchell O, Iqbal W, Lee SM, Foy KK, Lee KH, Moyer EJ, VanderArk A, Koeman JM, Ding W, Kalkat M, Spix NJ, Eagleson B, Pospisilik JA, Szabó PE, Bartolomei M, Vander Schaaf NA, Kang L, Wiseman AK, Jones PA, Krawczyk CM, Adams M, Porecha R, Chen BH, Shen H, Laird PW. 2022. DNA methylation dynamics and dysregulation delineated by high-throughput profiling in the mouse. Cell Genom 2(7):100144.
Jin SG, Meng Y, Johnson J, Szabó PE, Pfeifer GP. 2022. Concordance of hydrogen peroxide-induced 8-oxoguanine patterns with two cancer mutation signatures of upper GI tract tumors. Sci Adv 8(22): eabn3815.
Liao J, Zeng TB, Pierce N, Tran DA, Singh P, Mann JR, Szabó PE. 2021. Prenatal correction of IGF2 to rescue the growth phenotypes in mouse models of Beckwith-Wiedemann and Silver-Russell syndromes. Cell Rep 34(6):108729.
Zeng TB, Pierce N, Liao J, Singh P, Lau K, Zhou W, Szabó PE. 2021. EHMT2 suppresses the variation of transcriptional switches in the mouse embryo. PLoS Genet.
Zeng, TB, Pierce N, Szabó PE. 2021. H3K9 methyltransferase EHMT2/G9a controls ERVK-driven non-canonical imprinted genes. Epigenomics 13(16).
Liao J, Szabó PE. 2020. Maternal DOT1L is dispensable for mouse development. Sci Rep 10(1):20636.
Zeng TB, Szabó PE. 2020. Immunochemical detection of modified cytosine species in mammalian preimplantation embryos. Methods Mol Biol 2198:147–157.
Huang Z, Meng Y, Szabó PE, Kohli RM, Pfeifer GP. 2019. High resolution analysis of 5-hydroxymethylcytosine by TET-assisted bisulfite sequencing.Methods Mol Bio 2198:321–331.
Pfeifer GP, Szabó PE, Song J. 2019. Protein interactions at oxidized 5-methylcytosine bases. J Mol Biol.
Zeng TB, Han L, Pierce N, Pfeifer GP, Szabó PE. 2019. EHMT2 and SETDB1 protect the maternal pronucleus from 5mC oxidation. Proc Natl Acad Sci U S A. 116(22):10834-10841.
Pai S, Li P, Killinger B, Marshall L, Jia P, Liao J, Petronis A, Szabó P, Labrie V. 2019. Differential methylation of enhancer at IGF2 is associated with abnormal dopamine synthesis in major psychosis. Nat Comm.
Pfeifer GP, Szabó PE. 2018. Gene body profiles of 5-hydroxymethylcytosine: potential origin, function and use as a cancer biomarker. Epigenomics.
Jin SG, Zhang ZM, Dunwell TL, Harter MR, Wu X, Johnson J, Li Z, Liu J, Szabó PE, Lu Q, Xu GL, Song J, Pfeifer GP. 2016. Tet3 reads 5-carboxylcytosine through its CXXC domain and is a potential guardian against neurodegeneration. Cell Rep 14(3):493–505.
Szabó PE. 2015. Response to: the nature of evidence for and against epigenetic inheritance. Genome Biol 16:138
Iqbal K, Tran DA, Li AX, Warden C, Bai AY, Singh P, Wu X, Pfeifer GP, Szabó PE. 2015. Deleterious effects of endocrine disruptors are corrected in the mammalian germline by epigenome reprogramming. Genome Biol 16:59.
Hahn M, Szabó PE, Pfeifer GP. 2014. 5-hydroxymethylcytosine: a stable or transient DNA modification? Genomics 104(5):314–323.
Tran DA, Bai AY, Singh P, Wu X, Szabó PE. 2014. Characterization of the imprinting signature of mouse embryo fibroblasts by RNA deep sequencing. Nucleic Acids Res 42(3):1772–1783.
Liao J, He Y, Szabó PE. 2013. The Pou5f1 distal enhancer is sufficient to drive Pou5f1 promoter-EGFP expression in embryonic stem cells. Int J Dev Biol 57(9-10):725–729.
Singh P, Li AX, Tran, DA, Oates N, Kang E-R, Wu X, Szabó PE. 2013. De novo DNA methylation in the male germ line occurs by default but is excluded at sites of H3K4 methylation. Cell Rep 4(1):205–219.
Singh P, Szabó PE. 2012. Chromatin immunoprecipitation to characterize the epigenetic profiles of imprinted domains. Methods Mol Biol 925:159–172.
Singh P, Lee DH, Szabó PE. 2012. More than insulator: multiple roles of CTCF at the H19-Igf2 imprinted domain. Front Genet 3:214.
Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, Iqbal K, Shi YG, Deng Z, Szabó PE, Pfeifer GP, Li J, Xu GL. 2011. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 477(7366):606–610.
Abe M, Tsai SY, Jin SG, Pfeifer GP, Szabó PE. 2011. Sex-specific dynamics of global chromatin changes in fetal mouse germ cells. PLoS One 6(8):e23848.
Kang ER, Iqbal K, Tran DA, Rivas GE, Singh P, Pfeifer GP, Szabó PE. 2011. Effects of endocrine disruptors on imprinted gene expression in the mouse embryo. Epigenetics 6(7):937–950.
Iqbal K, Jin SG, Pfeifer GP*, Szabó PE*. 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci USA 108(9):3642–3647.
*Equally contributing authors
Singh P, Wu X, Lee DH, Li AX, Rauch TA, Pfeifer GP, Mann JR, Szabó PE. 2011. Chromosome-wide analysis of parental allele-specific chromatin and DNA methylation. Mol Cell Biol 31(8):1757–1770.
MCB Spotlight article
Lee DH, Tran D, Singh P, Oates N, Rivas GE, Larson GP, Pfeifer GP, Szabó PE. 2011. MIRA-SNuPE, a quantitative, multiplex method for measuring allele-specific DNA. Epigenetics 6(2):212–223.
Lee DH, Singh P, Tsai, SY, Oates, N, Spalla A, Spalla C, Brown L, Rivas G, Larson G, Rauch AT, Pfeifer GP, Szabó PE. 2010. CTCF-dependent chromatin bias constitutes transient epigenetic memory of the mother at the H19-Igf2 imprinting control region in prospermatogonia. PLoS Genet 6(11):e1001224.
Lee DH, Singh P, Tsark WM, Szabó PE. 2010. Complete biallelic insulation at the H19/Igf2 imprinting control region position results in fetal growth retardation and perinatal lethality. PLoS One 5(9):e12630.
Singh P, Cho J, Tsai SY, Rivas GE, Larson GP, Szabó PE. 2010. Coordinated allele specific histone acetylation at the differentially methylated regions of imprinted genes. Nucleic Acids Res 38(22):7974–7990.
Singh P, Han L, Rivas GE, Lee DH, Nicholson TB, Larson GP, Chen T, Szabó PE. 2010. Allele-specific H3K79 Di- versus trimethylation distinguishes opposite parental alleles at imprinted regions. Mol Cell Biol 30(11):2693–2707.
MCB Spotlight article
Han L, Lee DH, Szabó PE. 2008. CTCF is the master organizer of domain-wide allele-specific chromatin at the H19/Igf2 imprinted region. Mol Cell Biol 28(3):1124–1135.
Szabó PE, Han L, Hyo-Jung J, Mann JR. 2006. Mutagenesis in mice of nuclear hormone receptor binding sites in the Igf2/H19 imprinting control region. Cytogenet Genome Res 113(1-4):238–246.
Szabó PE, Pfeifer GP, Mann JR. 2004. Parent-of-origin-specific binding of nuclear hormone receptor complexes in the H19-Igf2 imprinting control region. Mol Cell Biol 24(11):4858–4868.
Szabó PE, Tang SH, Silva FJ, Tsark WM, Mann JR. 2004. Role of CTCF binding sites in the Igf2/H19 imprinting control region. Mol Cell Biol 24(11):4791–4800.
Szabó PE, Hübner K, Schöler H, and Mann JR. 2002. Allele-specific expression of imprinted genes in mouse migratory primordial germ cells. Mech Dev115(1-2):157–160.
Szabó PE, Tang SH, Reed MR, Silva FJ, Tsark WM, Mann JR. 2002. The chicken beta-globin insulator element conveys chromatin boundary activity but not imprinting at the Igf2 and H19 imprinted domain. Development 129(4):897–904.
Szabó PE, Pfeifer GP, Miao F, O’Connor TR, Mann JR. 2000. Improved in vivo dimethyl sulfate footprinting using AlkA protein: DNA-protein interactions at the mouse H19 gene promoter in primary embryo fibroblasts. Anal Biochem 283(1):112–116.
Szabó PE, Tang SH, Rentsendorj A, Pfeifer GP, Mann JR. 2000. Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr Biol 10(10):607–610.
Szabó PE, Pfeifer GP, Mann JR. 1998. Characterization of novel parent-specific epigenetic modifications upstream of the imprinted mouse H19 gene. Mol Cell Biol 18(11):6767–6776.
Szabó PE, Mann JR. 1996. Maternal and paternal genomes function independently in mouse ova in establishing expression of the imprinted genes Snrpn and Igf2r: no evidence for allelic trans-sensing and counting mechanisms. EMBO J 15(22):6018–6025.
Szabó PE, Mann JR. 1995. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev 9(24):3097–3108.
Szabó PE, Mann JR. 1995. Biallelic expression of imprinted genes in the mouse germline: implications for erasure, establishment, and mechanisms of genomic imprinting. Genes Dev 9(15):1857–1868.
Szabó P, Moitra J, Rencendorj A, Rákhely G, Rauch T, Kiss I. 1995. Identification of a nuclear factor-I family protein-binding site in the silencer region of the cartilage matrix protein gene. J Biol Chem 270(17):10212–10221.
Szabó P, Mann JR. 1994. Expression and methylation of imprinted genes during in vitro differentiation of mouse parthenogenetic and androgenetic embryonic stem cell lines. Development 120(6):1651–1660.
Kiss I, Bösze Z, Szabó P, Altanchimeg R, Barta E, Deák F. 1990. Identification of positive and negative regulatory regions controlling expression of the cartilage matrix protein gene. Mol Cell Biol 10(5):2432–2436.


Brandon Estrem, Ph.D.
Postdoctoral Fellow, Pfeifer & Szabó Laboratories

Ji Liao, Ph.D.
Senior Lab Manager, Department of Epigenetics

Amy Nuffesse
Senior Administrative Assistant II, Department of Epigenetics


